Reasearch topics
The team's research focuses on understanding the structural and functional organisation of the nucleolus and 45S ribosomal RNA (45S rDNA) genes in plants. In all eukaryotic organisms, RNA pol I transcription of the 45S rDNA and processing of the 45S pre-rRNA is the keystone of ribosome biogenesis and nucleolus assembly. In particular, rRNAs are modified at specific sites (methylation of sugars and/or bases, pseudouridylation, acetylation, etc.). These rRNA cleavages (via the major/ITS-1 first or minor 5'ETS first pathway) and modifications are conserved in higher eukaryotic cells and necessary for the stability of the ribosome structure and the accuracy and fine-tuning of translation. In the team we tackled these issues during plant growth and development and in response to cellular and environmental conditions.
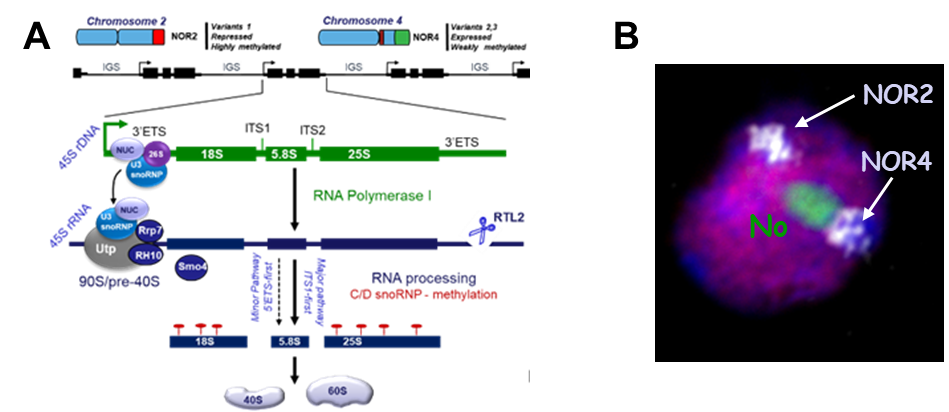
Figure 1. The ribosomal rDNA genes (45S rDNA) are organised in tandem repeats in nucleolar organiser region (NOR) in chromosomes 2 and 4 (NOR2 and NOR). The NOR2 contain 45S rDNA variants type 1 while the NOR4 contain 45S rDNA variants type 2 and 3. NOR2 and NOR4 are usually silent or transcriptionally active, respectively. Each 45S rDNA unit encodes the 18S, 5,8S and 25S rRNAs separated by external (5’and 3’ETS) and internal (ITS-1 and -2) transcribed spacers. The 25S, 5.8S, 5S (transcribed by RNA pol III) the large (LSU) and 18S rRNAs form small (SSU) subunits of the ribosome. The LSU acts as a ribozyme, catalysing the formation of peptide bonds, while the SSU houses the decoding or translation centre. In all eukaryotic cells, rRNAs are subject to nucleotide modifications, including 2’-O-methylation driven by small C/D snoRNP complexes. The assembly of 40S and 60S particles initiates on nascent pre-rRNA, and requires hundreds of factors, including, Nucleolin-U3snoRNP complex, RTL2, and Rrp7, Smo4; Smo2 and RNA helicase RH10. Theses factor and modifications were studied in the last past years. B) Visualization of NORs by DNA FISH (white) and of the nucleolus by immunolocalization of NUC1 (green). Euchromatin is labeled using anti-H3K4me3 antibodies (purple).
Keywords: nucléole, ribosome, rDNA, rRNA, modification de RNA, snoRNA
Funded projects:
- ANR MetRibo (05/2021-10/2025): Tackling rRNA Methylation: Molecular Bases and Ribosome Translational Impact: Partners: J. Sáez-Vásquez (CNRS/UPVD, Perpignan), I. Motorin (U. de Lorraine, Strasbourg); C. Gaspin (INRA, Toulouse).
- ANR RoxRNase (04/2021-09/2025): Redox regulation in small RNA-mediated responses to biotic and abiotic stresses. Partners J-P. Reichheld (CNRS/UPVD, Perpignan); J. Sáez-Vásquez (CNRS/UPVD, Perpignan), H. Vaucheret (INRA, Versailles. CES12 Genetic, Genomic and RNA.
- ANR RiboStress (01/2018-12/2021). Genetic, epigenetic and protein-based crosstalks between nucleolus functional reorganization and response to stress. Partners: J. Sáez-Vásquez (CNRS/UPVD, Perpignan) F. Barneche (ENS, Paris), C. Carapito (CNRS/U de Strasbourg) and F. Le Galliard (CEREEP-ECOTRON IDF).
Members
Past membres
- Eduardo Muñoz Díaz
- Charlotte Montacié
- Erwen Lemesle (CDD IE, ANR RoxRNAse)
- Sara-Alina Neumann (CDD/Post doc Chercheur, MetRibo)
- Tommy Darrière (CDD/Post doc Chercheur, RiboStress)
Distinction
NASA Group Achievements Award 2019 : https://www.insb.cnrs.fr/fr/cnrsinfo/le-nasa-group-achievement-award-2019-attribue-m-julio-saez-vasquezPublication par collection
- Articles
-
- Communications
-
- Thèse et HDR
-