Team PI : MIROUZE Marie et PANAUD Olivier
The team is studying several plant models, including the model plant Arabidopsis thaliana, cultivated rice (Oryza sativa), beech (Fagus sylvatica) and Dittrichia viscosa, a source of biopesticide. The team specialises in developing protocols for using Nanopore long reads in genomics and transcriptomics, as well as developing dedicated bioinformatics tools. Lastly, our team is involved in participatory science and training programmes on circular extrachromosomal DNA (eccDNA) and Nanopore technology (e.g. science in high schools and workshops).
We are developing the following research themes:
(1) The mechanisms of genomic biodiversity
(2) The contribution of genomics to sustainability science and the fight against climate change
(3) Development with private partners
(1) Mechanisms of genomic biodiversity
- Epigenome and genome stability in Arabidopsis (coordinated by Marie Mirouze).
As part of the ANR 'CropCircle' project (collaboration with Dr Etienne Bucher, Agroscope, Switzerland), we are analysing the production dynamics of circular extrachromosomal DNA (eccDNA) under stressful environmental conditions, as well as its impact on genome stability in Arabidopsis thaliana.
- Introgressions and grain width in rice (coordinated by Marie-Christine Carpentier):
We are developing original markers based on transposable element insertion polymorphisms (TIPS) and using association genomics approaches (GWAS) to identify loci of interest in rice grain width, in collaboration with our colleagues from Taiwan (Academia Sinica).
(2) The science of sustainability
- The beech genome in the face of climate change (coordinated by Olivier Panaud)
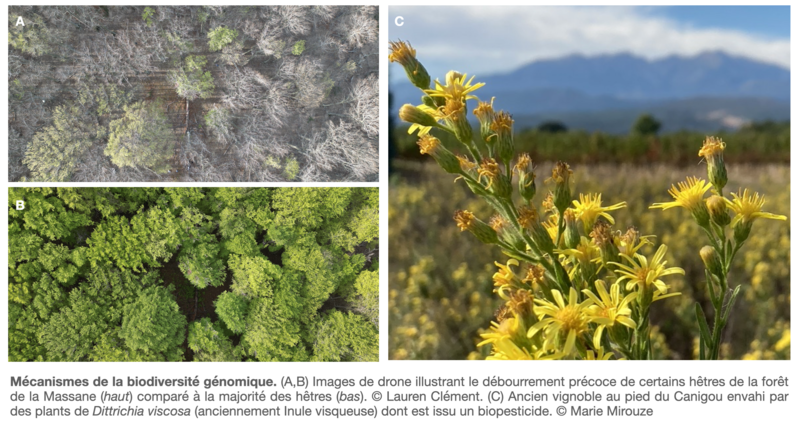
In order to understand the links between genome dynamics and the adaptive potential of beech populations, we are analysing the genomic diversity of beech trees, with a particular focus on those in Mediterranean forests (specifically the Massane Nature Reserve).
- Dynamics of the rice genome in Mali (coordinated by Marie Mirouze).
In partnership with Pr Ousmane Koita's laboratory in Bamako, Mali, we are developing genomic and transcriptomic approaches to improve our understanding of rice's resistance to multiple pathogen infections.
(3) Development with the private sector
- Inulin and biopesticides (MANGO/AKINAO project):
Biopesticides offer an interesting alternative for reducing agricultural inputs. In partnership with the AKINAO company (www.akinao-lab.com), we are characterising the genome of slimy inula (Dittrichia viscosa) to establish this plant as a model for biopesticide production.
- The contribution of eccDNA to cancer detection (MANGO/ACOBIOM project).
Cancer cells produce large quantities of eccDNA. In partnership with ACOBIOM, we are using our expertise in eccDNA to develop early markers for various cancers.

Research topics
Ever since Charles Darwin's pioneering work, we have known that natural selection is based on pre-existing or induced variations. The MANGO team is interested in the molecular mechanisms underlying these variations within and between individuals. This research aims at understanding the role of genome dynamics in plants adapting to climate change. Far from being a fixed entity, the genome contains transposable elements that can generate variation by moving within chromosomes. These structural variations (notably caused by transposable elements, but not only) may result from environmental stress or epigenetic deregulation, impacting gene expression and adaptation.The team is studying several plant models, including the model plant Arabidopsis thaliana, cultivated rice (Oryza sativa), beech (Fagus sylvatica) and Dittrichia viscosa, a source of biopesticide. The team specialises in developing protocols for using Nanopore long reads in genomics and transcriptomics, as well as developing dedicated bioinformatics tools. Lastly, our team is involved in participatory science and training programmes on circular extrachromosomal DNA (eccDNA) and Nanopore technology (e.g. science in high schools and workshops).
We are developing the following research themes:
(1) The mechanisms of genomic biodiversity
(2) The contribution of genomics to sustainability science and the fight against climate change
(3) Development with private partners
(1) Mechanisms of genomic biodiversity
- Epigenome and genome stability in Arabidopsis (coordinated by Marie Mirouze).
As part of the ANR 'CropCircle' project (collaboration with Dr Etienne Bucher, Agroscope, Switzerland), we are analysing the production dynamics of circular extrachromosomal DNA (eccDNA) under stressful environmental conditions, as well as its impact on genome stability in Arabidopsis thaliana.
- Introgressions and grain width in rice (coordinated by Marie-Christine Carpentier):
We are developing original markers based on transposable element insertion polymorphisms (TIPS) and using association genomics approaches (GWAS) to identify loci of interest in rice grain width, in collaboration with our colleagues from Taiwan (Academia Sinica).
(2) The science of sustainability
- The beech genome in the face of climate change (coordinated by Olivier Panaud)
In order to understand the links between genome dynamics and the adaptive potential of beech populations, we are analysing the genomic diversity of beech trees, with a particular focus on those in Mediterranean forests (specifically the Massane Nature Reserve).
- Dynamics of the rice genome in Mali (coordinated by Marie Mirouze).
In partnership with Pr Ousmane Koita's laboratory in Bamako, Mali, we are developing genomic and transcriptomic approaches to improve our understanding of rice's resistance to multiple pathogen infections.
(3) Development with the private sector
- Inulin and biopesticides (MANGO/AKINAO project):
Biopesticides offer an interesting alternative for reducing agricultural inputs. In partnership with the AKINAO company (www.akinao-lab.com), we are characterising the genome of slimy inula (Dittrichia viscosa) to establish this plant as a model for biopesticide production.
- The contribution of eccDNA to cancer detection (MANGO/ACOBIOM project).
Cancer cells produce large quantities of eccDNA. In partnership with ACOBIOM, we are using our expertise in eccDNA to develop early markers for various cancers.
Key words : genome, transposable elements, extrachromosomal DNA, structural variants, epigenome
Financed projects
- ANR FAGRESCUE (2025-2028): Une approche transdisciplinaire impliquant l'intelligence artificielle, la génomique et la modélisation écologique pour améliorer la résilience des hêtraies dans le contexte du changement climatique. LGDP leader (Olivier Panaud), Partenaires: ONERA (DOTA), CEFE, INRAE.
- Projet Innovant Fondation UPVD (Appel à projets 2024) : PI Marie Mirouze, partenaire société ACOBIOM.
- Projet Bonus Qualité Recherche (BQR) UPVD (2024) - Impact fonctionnel des variations structurales chez le riz sauvage Oryza australiensis. PI : Marie-Christine Carpentier.
- Projet Innovation Labex TULIP DROPHENO : Combiner l'imagerie par drone, l'intelligence artificielle et l'écologie de terrain pour concevoir une stratégie de phénotypage à haut débit des forêts de hêtres. Partenaire: Bionomeex
- Projet pilote BioDivOc FAGADAPT (Appel à projets 2024, appel à projets 2022). Adaptation des forêts tempérées au changement climatique. Une étude de cas combinant génomique et écologie pour suivre l’adaptation à court terme de la hêtraie dans la réserve de la Massane. LGDP leader (Olivier Panaud), Partenaires: CEFE, BIOM, Réserve Naturelle de la Massane.
- Projet Innovation Labex TULIP (2021) : Etudier la diversité génétique et la production de composés biofongicides à partir de différentes populations de Dittrichia viscosa du pourtour méditerranéen. PI Fred Pontvianne, Partenaire M Mirouze.
- ANR PRCI CropCircle (2021-2025) Mobilome, adaptation et stabilité du génome chez les plantes cultivées. PI Marie Mirouze (FR) et Etienne Bucher (CH)
- Projet Central Labex TULIP (2020) : Comprendre le rôle du lierre commun (Hedera helix) dans les transferts horizontaux entre arbres. PI Moaine El Baidouri.
- EpiDiverse (2017-2021) Projet Européen MCSA European Training Network grant, EU H2020 Marie Skłodowska-Curie No 764965 EpiDiverse
- ANR JCJC EXTRACHROM (2014-2017). Contrôle épigénétique des formes extrachromosomiques des éléments transposables chez les plantes. PI Marie Mirouze
Members
Past members
- Abirami Soundiramourtty (2021 - 2024)
- Panpan Zhang (2020 - 2023)
- Emilie Aubin (2019 - 2022)
- Sophie Lanciano (2014 - 2017)
Publications
- Articles
-
- Communications
-
- Thèse et HDR
-
Updated : June 23, 2025

