Our laboratory studies the evolutionary processes involved in plant genome plasticity as well as the molecular mechanisms that regulate gene expression, primarily under stress. We use approaches from genomics, bioinformatics, theoretical biology and modeling, genetics, biochemistry, and cell and molecular biology.
The laboratory is organised in teams :
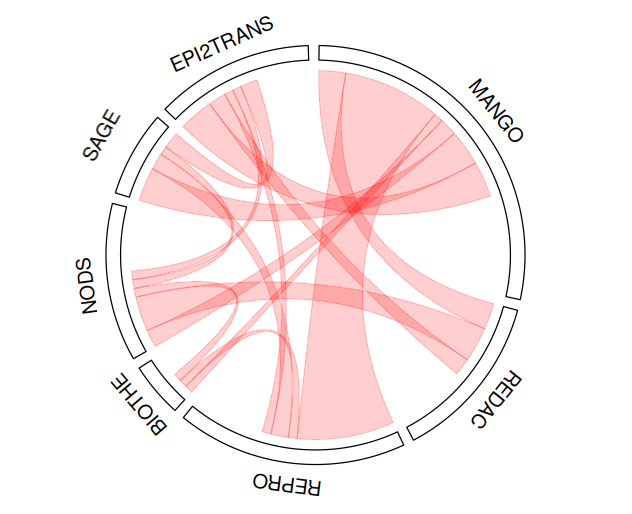
Illustration of collaborations between teams by representing the number of articles published jointly over the years 2020 - 2024 :
The list of funded projects supported by the LGDP is accessible : list of funded projects.
The laboratory is organised in teams :
| 2020 - 2024 | 2025 - 2029 | Team |
| SipTRiP | REPRO | Post-transcriptional regulation of gene expression in Arabidopsis |
| GRRIP | SAGE | Silencing and gene regulation in photosynthetic organisms |
| MANGO | MANGO | Adaptation mechanisms and genomics |
| REDAC | REDAC | Redox, Development and Adaptation to Constraints |
| NoDS | NODS | Nucleolus: development and stress response |
| MEAC | EPI2TRANS | Epigenetic regulations of transcription and transposons |
| BT | BIOTHE | Theoretical biology |
| GENOMIX | Genomics genetic exchanges adaptation & evolution |

The list of funded projects supported by the LGDP is accessible : list of funded projects.
Updated : June 23, 2025

