Research topics
Most of the genetic information encoded by genomic DNA is first transcribed as messenger RNA (mRNA), which is then translated by ribosomes to produce proteins that perform numerous cellular functions. Decades of active research have revealed that various regulatory mechanisms operating at different levels (transcriptional, post-transcriptional or translational/post-translational) in this flow of genetic information are used in combination to control and modulate gene expression and the activity of their final protein products. Among the means available to the plant cell to regulate gene expression, we are interested in mechanisms involving epigenetic modification of chromatin, epitranscriptomic modification of mRNA bases, RNA-binding protein/RBP transactions, and small RNA-guided gene regulation by Argonaute proteins.Epitranscriptomic-based regulation in plants.
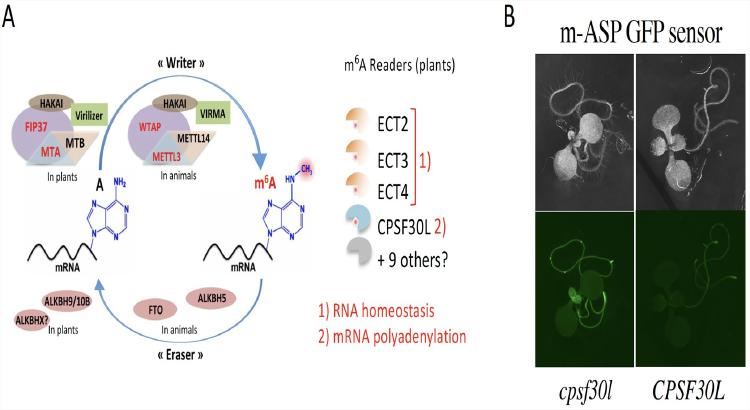
Fig.1:Mechanisms A) and in planta visualization B) of m6A-assisted polyadenylation pathway.
mRNA fate in eukaryotes is controlled by chemical modification of nucleotides, in a mechanism known as “epitranscriptomics”. m6A has emerged as a prevalent mRNA modification, and the current idea is that the m6A mark is co-transcriptionally deposited by a conserved writer complex (MTC), and then primarily controls mRNA fate by recruiting readers that belong to the YTH domain-containing protein family (also known as ECT in plants). Mutation in any of these components prevents m6A methylation and has impacts on many biological processes, including development, stress adaptation, hormone response, antiviral response and cancer.
Our team has uncovered an m6A-assisted polyadenylation/m-ASP mechanism in Arabidopsis, that ensures transcriptome fidelity through the use of a specific YTH-type reader protein (CPSF30L) (Fig1A). Interestingly, a similar pathway also exists in the Apicomplexan parasite Toxoplasma gondii, as shown in collaboration with Dr. M.-A. Hakimi’s team (IAB, Grenoble).
We are currently interested in identifying new players in the m-ASP and m6A pathways in plants. For this purpose, we have generated a GFP-based m-ASP reporter sensor (Fig.1B) that is currently used in a forward genetic screen as a first step to characterize candidates. They will be analyzed by a large panel of biochemical and molecular approaches (IP-MS, CLNIP-MS, ChIP-seq, Nanopore DRS....).
Roles of RNA-binding proteins in plants development.
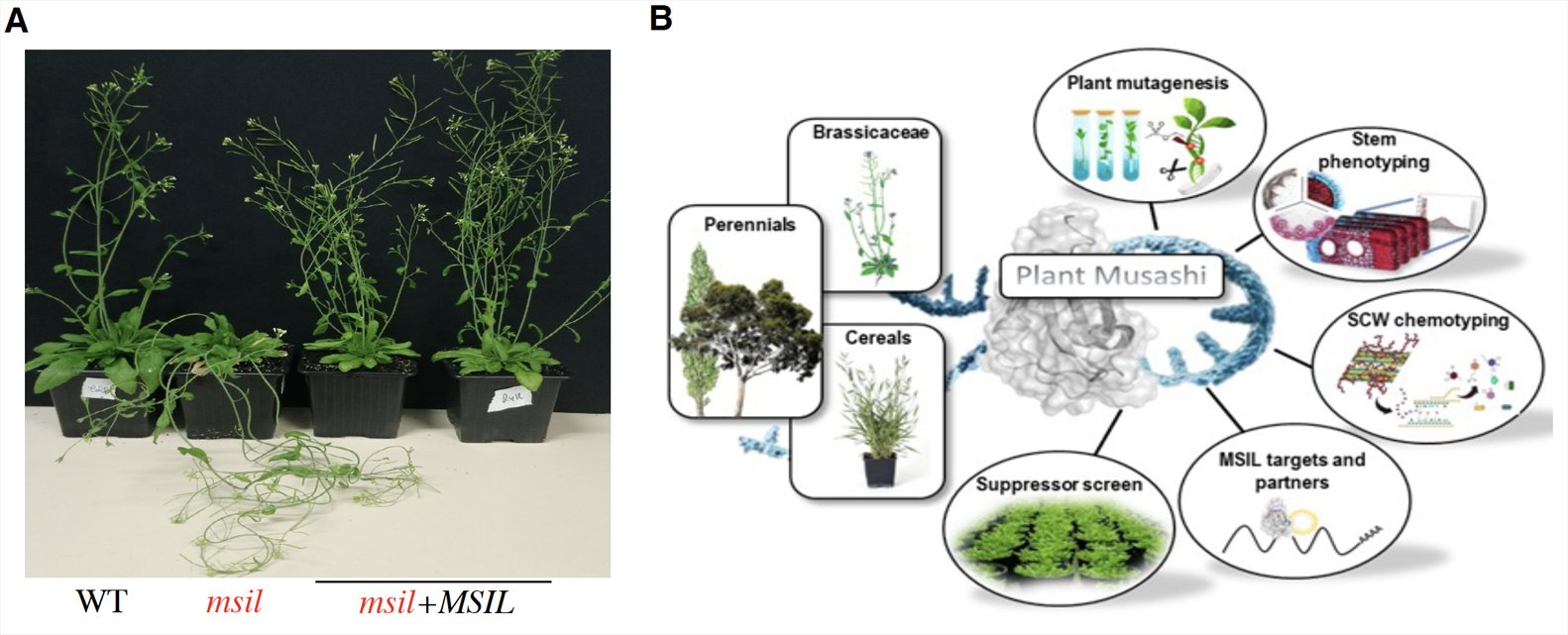
Fig.2: A) secondary cell wall defect in msil mutants and B) Integrative analysis of Musashi-like RBP in plants.
RNA-binding proteins (RBPs) are essential posttranscriptional modulators of gene expression across all kingdoms of life. However, although a large cohort of RBPs exists in plants, there is little information about RBP-mediated posttranscriptional regulation of SCW biosynthetic genes. We reported that plant RNA-binding proteins homologous to the animal translational regulator Musashi (MSIL) control various aspects of Arabidopsis development, including the formation of secondary cell wall/SCW in the inflorescence stem. This result points to a new mechanism for control of secondary cell wall synthesis, linking post-transcriptional/translational control to the regulation of SCW biosynthesis genes. We are currently applying a unique combination of molecular, biochemical, genetic and chemo-typing approaches to shed light on the function of MSIL function in Arabidopsis, but also in tree and cereal plant models that present SCWs with different chemical structures.
Roles and regulation of Argonaute effector proteins in RNA silencing in plants.
Sessile organisms such as plants are particularly vulnerable in a context of globalization and climate change. Over the course of their evolution, plants have acquired specialized and sophisticated mechanisms of gene regulation by small RNA to ensure their development and adapt to these constraints. These regulatory pathways, known as RNA silencing, are widely conserved in living organisms, and the understanding of their mode of action was rewarded with two Nobel Prizes in 2006 and 2024 (Fig.3A). RNA silencing pathways often use an Argonaute/AGO protein effector which binds to the small RNA guide and silence complementary DNA/mRNA targets.
Plants are distinguished by a large number of AGO proteins, which can be classified into two main groups: the AGO1 clade, whose members bind 21-nt long miRNAs/siRNAs and play a major role in plant development and response to the environment through post-transcriptional gene regulation (Fig.3B), and the AGO4 clade, whose members bind 24 nt-long siRNA and silence genomic repeats at a transcriptional level. Deciphering the mode of action of this superfamily of effectors is therefore crucial to understanding RNA silencing mechanisms.
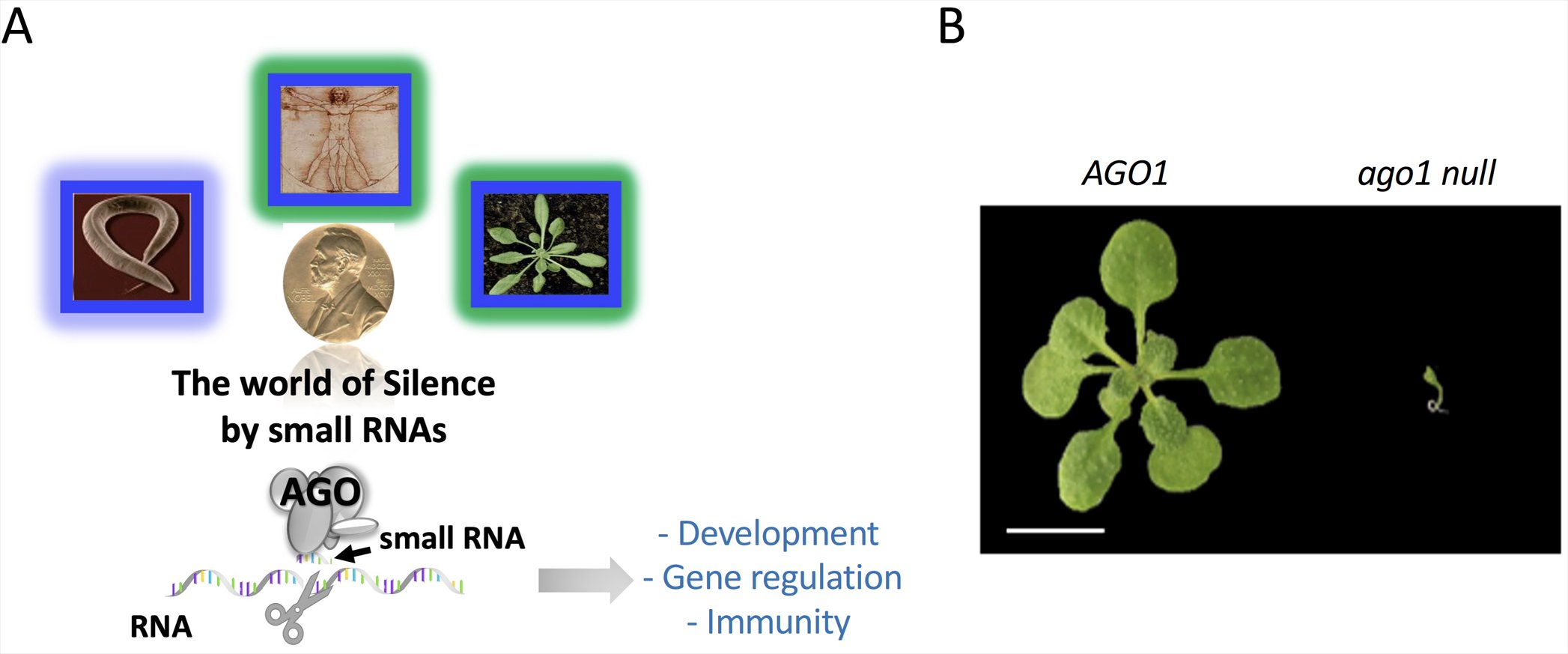
Fig.3: RNA silencing is a widely conserved sequence-specific lever for silencing gene expression through the action of small RNA-guided Argonaute proteins on their complementary targets (Post-transcriptional Gene Silencing shown here with mRNA as target) A), and developmental phenotype of the ago1 null mutant in Arabidopsis B).
We are currently following two different approaches to assess AGO protein activity in plants: the first one is based on a non-biased bioinformatic screen while the second focuses on the fine-tuned regulation of one major Argonaute protein, AGO1.
1) The WG (tryptophan-glycine) AGO hook motif is a conserved epitope used by proteins to recruit AGO partners in various RNA silencing pathways. We have previously developed a bioinformatic approach to assess the genome-wide distribution of AGO hook motif in proteomes, identifying potential AGO-anchor proteins in both plant, animal and associated pathogens proteomes. We are currently following up the functional analysis of AGO-anchor candidates identified, in the hope of revealing new RNA silencing activities in plants.
2) AGO1 activity is tightly controlled at several levels, but its regulation at the post-translational level remains poorly understood in plants, unlike its animal counterparts. Indeed, post-translational modifications (PTMs) are a particularly interesting mode of regulation, given the fundamental biological processes regulated by AGO1. Among PTMs, methyl groups can be added to arginine side chains of protein by the action of arginine/R methyltransferases/PRMTs (R-met), thus modulating their fate (turnover, localization) and/or their function (folding, activity, partner network). Our work on the model plant Arabidopsis thaliana has shown that AGO1 is methylated by different members of the PRMT protein family, revealing a unique double R-met signature within the AGO/PIWI protein superfamily, and even more generally in animal and plant proteins. We are currently developing a project aimed at understanding the biological and functional relevance of these R-met modifications for AGO1 action and, more broadly for RNA silencing pathways in plants. We are currently using a large panel of approaches such as plant phenotyping in normal or stress conditions and biochemical or molecular techniques more particularly adapted to smallRNA studies (stem-loop QPCR, smallRNA-Seq...).
Key words : epigenetics, epitranscriptomics, transcription, Argonaute, siRNA, m6A, Musashi, development, RBP, Arginine methylation, adaptation to biotic and abiotic stresses.
Funded projects
- ANR MUSAWALL (2023-2028). Mechanisms and contributions of Musashi-mediated control in cell wall polymers synthesis in plants. Coordinator: Thierry Lagrange (LGDP, CNRS), Partners: Richard Sibout (BIA, CNRS/INRAe); Fabien Mounet (LRSV, UT3).
- ANR ALGALVIRUS (2017-2022). Mechanisms and contributions of Musashi-mediated control in cell wall polymers synthesis in plants. Coordinator: Thierry Lagrange (LGDP, CNRS), Partners: Richard Sibout (BIA, CNRS/INRAe); Fabien Mounet (LRSV, UT3).
- UPVD University Grant (Bonus Qualité Recherche/BQR) (2024-25) - The R-met pathway, a global regulator of immunity in plants. PI : Jacinthe Azevedo-Favory.
- Central Project Labex TULIP (2022) – Functional analysis of Musashi-type proteins in Arabidopsis thaliana. PI : Natacha Bies-Etheve.
- Central Project Labex TULIP (2020) - Analysis of the impact of the PRMT5 enzyme on the AGO1 protein in A. thaliana. PI : Jacinthe Azevedo-favory.
Members
Past members
- Clément Barré-Villeneuve
- Alicia Kairouani
Publications
- Articles
-
- Communications
-
- Thèse et HDR
-

