Post-transcriptional regulation of gene expression in Arabidopsis
Research topics
The timely reprogramming of gene expression in response to internal and external cues is essential to eukaryote development and acclimation to changing environments. The reprogramming of gene expression involves many steps, from transcription and translation to post-translational modifications and degradation of RNA and proteins. To obtain an integrated view of the mechanisms regulating gene expression, it is essential to obtain information on all these steps and the way they influence each other. Our current understanding of how plant gene expression is regulated concerns mainly the gene transcription step. Our main objective is to increase our knowledge on key post-transcriptional steps of plant gene expression focusing on mRNA stability and translation, particularly in stress conditions and during development.
A) B)
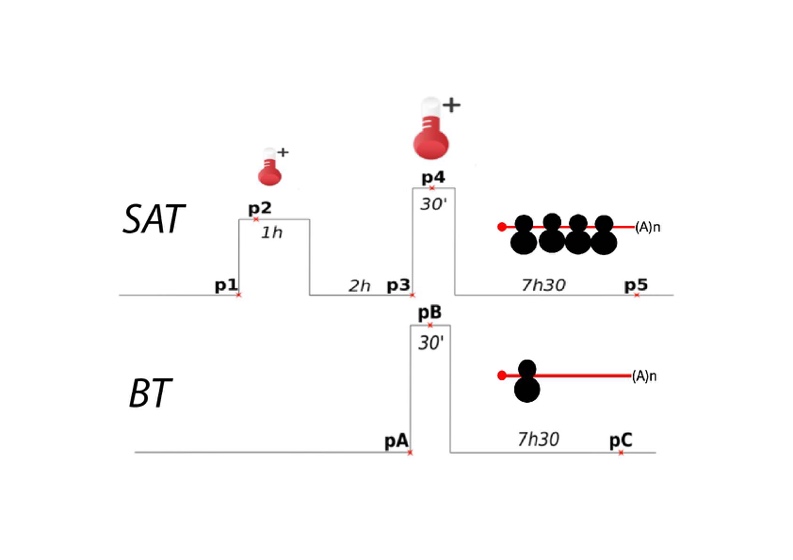

A) How plant response to intermittent heat stress involves modulation of mRNA translation efficiency. Acquired thermotolerance (also known as priming) is the ability of cells or organisms to better survive an acute heat stress if it is preceded by a milder one. In plants, acquired thermotolerance has been studied mainly at the transcriptional level, including recent descriptions of sophisticated regulatory circuits that are essential for this learning capacity. In this project, we study the involvement of polysome-related processes (translation and cotranslational mRNA decay) in plant thermotolerance using two heat stress regimes with and without a priming event.
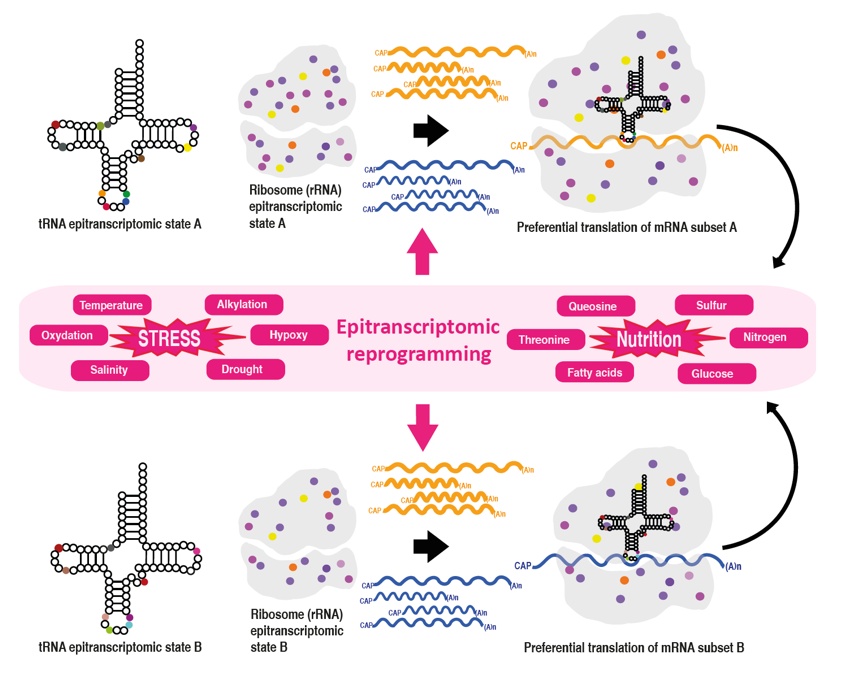
B) How variation in tRNA epitranscriptomic status impact translation and contribute to plant adaptation to environmental challenges. Recently, mRNA chemical (i.e., epitranscriptomic) modifications were shown to play a key role in regulating gene expression. In contrast, tRNA chemical modifications, although critical for optimal function of the translational apparatus, and much more diverse and quantitatively important compared to mRNA modifications, were until recently considered as mainly static chemical decorations. In this project, we challenge this view and test the hypothesis that plant tRNA modifications dynamically respond to various cell and environmental conditions and contribute to development and stress responses.
Key words: Translation, heat stress, tRNA, epitranscriptomic, ribosome, Arabidopsis
Funded projects
- ANR Heat-Adapt (2014-2019) Post-transcriptional regulation of heat stress response in plants. Partners: J.M. Deragon (CNRS/UPVD, Perpignan), Y.Y. Charng (ABRC, Taipei)
- ANR 3'ModRN (2016-2021): Modifications of mRNA 3' ends by nucleotide addition: impact on translation and mRNA degradation in Arabidopsis thaliana. Partners: D. Gagliardi (CNRS, Strasbourg), C. Bousquet-Antonelli (CNRS/UPVD, Perpignan)
- ANR Heat-EpiRNA (2018-2022): Modifications of mRNA 3' ends by nucleotide addition: impact on translation and mRNA degradation in Arabidopsis thaliana. Partners: D. Gagliardi (CNRS, Strasbourg), C. Bousquet-Antonelli (CNRS/UPVD, Perpignan)
- ANR Matilda (2021-2025): Molecular mechanism and physiological significance of 5'-3' co-translational mRNA degradation in Arabidopsis. Partners: R. Merret (CNRS/UPVD, Perpignan), D. Gagliardi (CNRS, Strasbourg)
- ANR Epi-tRNA (2022-2026): Dynamics of the tRNA epitranscriptome: Phenotypic and molecular consequences on plant development and stress response. Partners: JM Deragon (CNRS/UPVD, Perpignan), L. Drouard-Maréchal (CNRS, Strasbourg), C. Hirtz (CHU, Montpellier)
Members
Past members
- Rémy Merret
- Cécile Bousquet-Antonelli
- Arnaud Dannfald
- Jérémy Scutenaire
- Celso Gaspar Litholdo
Publications
- Articles
-
- Communications
-
- Thèse et HDR
-









